Postgraduates of Biological Chemistry, Biophysics and BioengineeringSociety
3D image processing and particle tracking in low contrast live-cell images
Abstract
A true view of the function of molecular machines in cells involves live-cell imaging of molecular probes in 3 dimensions (3D) at high temporal resolution. While advanced fluorescence microscopes are now able to routinely capture 3D data stacks in time lapse mode to detect behaviour of multiple molecular species, the ability to analyse such images lags behind that to generate the primary data. There is now a growing interest and urgent need to develop sophisticated methods for analysing 3D time-lapse data sets in order to characterise the relative behaviour of the components of molecular machines and their dynamics. Our ability to segment and quantify single molecule sub-cellular components is currently hampered by the limitations of computational tools available for tracking nanometre scale particles in low contrast and low Signal to Noise Ratio (SNR) environments. In this project, I hope to explore new image processing approaches and methods for noise reduction, particle detection, particle tracking and statistic analysis for applications in the important area of molecular biology. Whilst developing these new approaches the people who will actually being using these tools will be kept in mind, as such they will be simple to implement and use for people of all computational skill levels.
Researchers
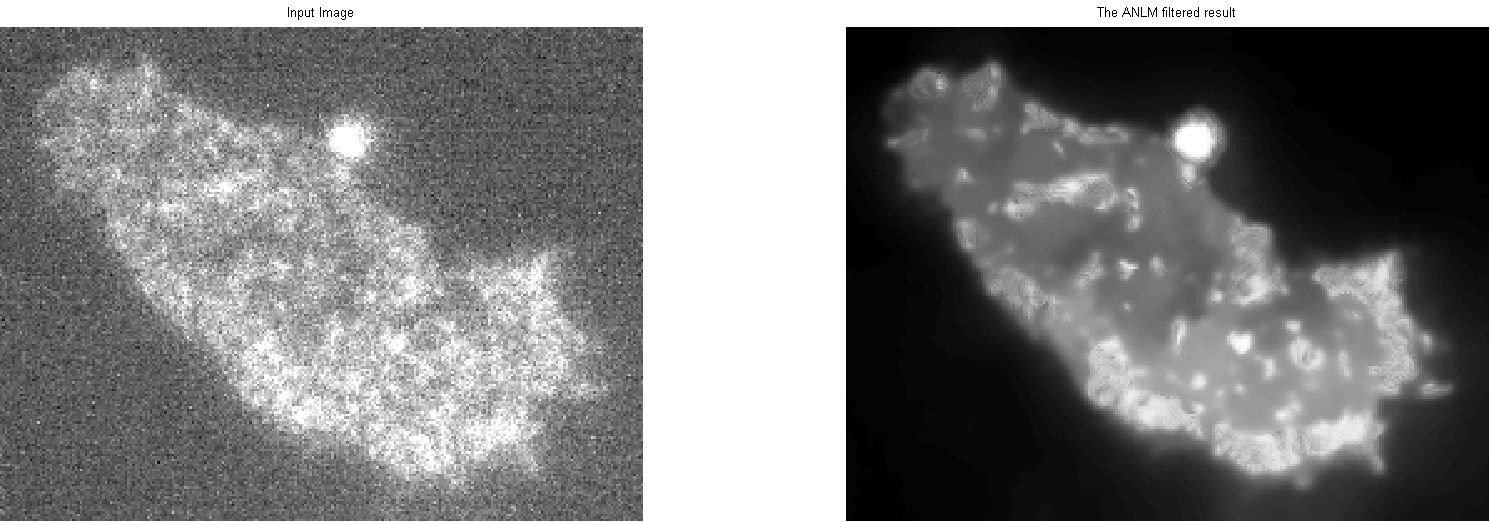
Figure 1: De-noised Image. Left: Original input “noisy” image. Right: De-Noised output image filtered using ANLM.
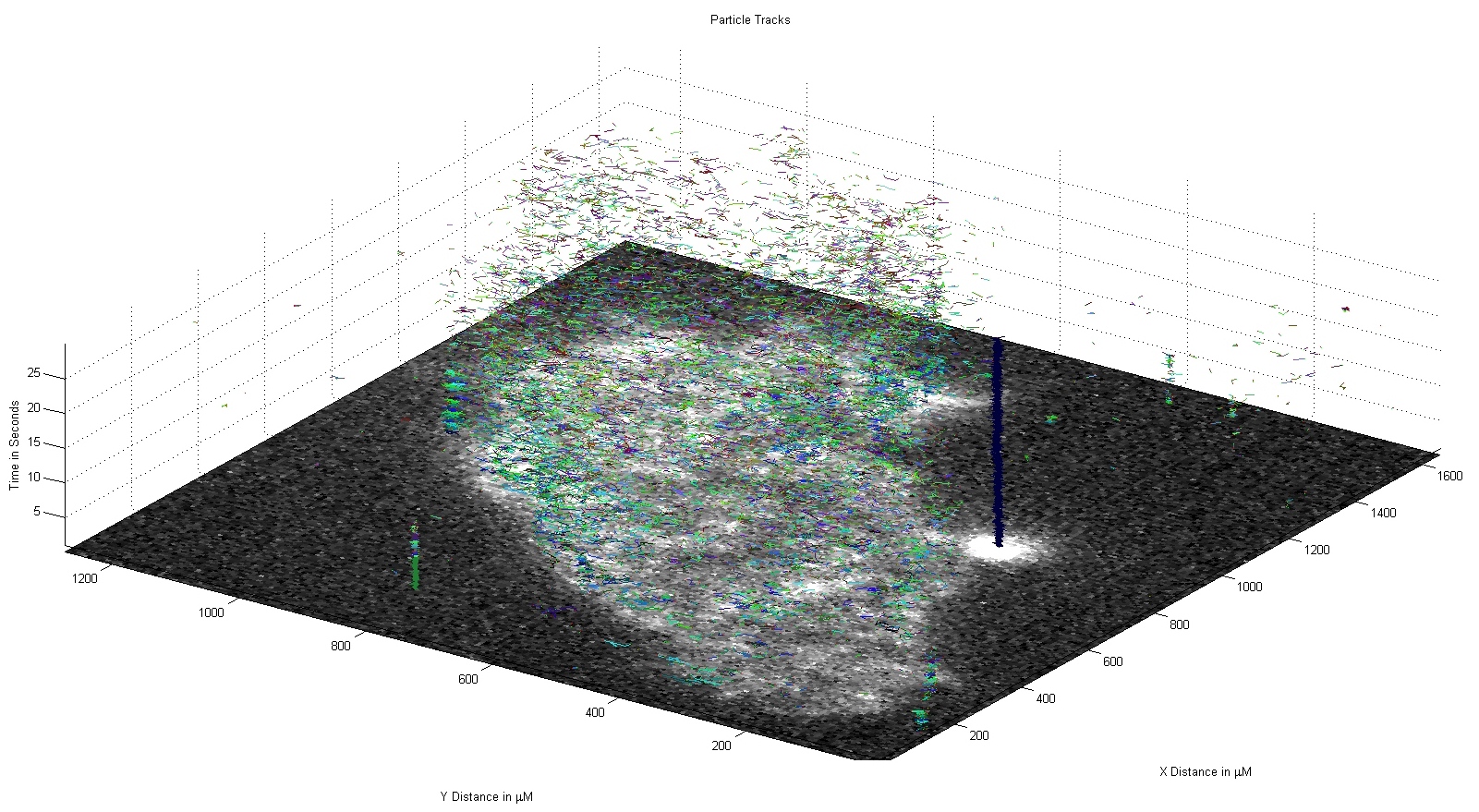
Figure 2: Tracked particles in a 2-dimensional space over time.